STA 235H - Model Selection I:
Bias vs Variance, Cross-Validation, and Stepwise
Fall 2023
McCombs School of Business, UT Austin
Announcements
Re-grading for homework 3 available until this Thursday.
- Please check the rubric and based on that ask for a specific re-grade.
Announcements
Re-grading for homework 3 available until this Thursday.
- Please check the rubric and based on that ask for a specific re-grade.
Think of assignment drop as an insurance policy.
- Start assignments with enough time if you already think you used your drop.
Announcements
Re-grading for homework 3 available until this Thursday.
- Please check the rubric and based on that ask for a specific re-grade.
Think of assignment drop as an insurance policy.
- Start assignments with enough time if you already think you used your drop.
Grades for the midterm will be posted on Tuesday.
Importance of completing assignments (e.g. practice quiz, JITTs).
Final exam will have limited notes.
Announcements
Re-grading for homework 3 available until this Thursday.
- Please check the rubric and based on that ask for a specific re-grade.
Think of assignment drop as an insurance policy.
- Start assignments with enough time if you already think you used your drop.
Grades for the midterm will be posted on Tuesday.
Importance of completing assignments (e.g. practice quiz, JITTs).
Final exam will have limited notes.
Start of a completely new chapter
- If you struggled with causal inference, doesn't mean that you can't do very well in this second part.
Last class

Finished with causal inference, discussing regression discontinuity designs
We will review the JITT (slides will be posted tomorrow)
Importance of doing the coding exercises
JITT 9: Regression discontinuity design
- RDD allows us to compare people exactly at the cutoff if they were treated vs not treated, and estimate a Local Average Treatment Effect (LATE) for those units.
JITT 9: Regression discontinuity design
RDD allows us to compare people exactly at the cutoff if they were treated vs not treated, and estimate a Local Average Treatment Effect (LATE) for those units.
In the example for the JITT, the treatment is being legally able to drink (and the control is not being legally able to drink).
JITT 9: Regression discontinuity design
RDD allows us to compare people exactly at the cutoff if they were treated vs not treated, and estimate a Local Average Treatment Effect (LATE) for those units.
In the example for the JITT, the treatment is being legally able to drink (and the control is not being legally able to drink).
The code you had to run is:
summary(rdrobust(mlda$all, mlda$r, c = 0))In this case, remember that
allis our outcome (total number of arrests),ris our centered running variable (age minus the cutoff), andc = 0is our cutoff (remember thatris centered around 0, so the cutoff is 0 and not 7670).You have to look at the coefficient in the table (
Conventional)... and remember to also look at the p-value!
JITT 9: Regression discontinuity design
RDD allows us to compare people exactly at the cutoff if they were treated vs not treated, and estimate a Local Average Treatment Effect (LATE) for those units.
In the example for the JITT, the treatment is being legally able to drink (and the control is not being legally able to drink).
The code you had to run is:
summary(rdrobust(mlda$all, mlda$r, c = 0))In this case, remember that
allis our outcome (total number of arrests),ris our centered running variable (age minus the cutoff), andc = 0is our cutoff (remember thatris centered around 0, so the cutoff is 0 and not 7670).You have to look at the coefficient in the table (
Conventional)... and remember to also look at the p-value!
"On average, for individuals with exactly 21 years of age, being legally able to drink increases the total number of arrests by 409.1, compared to not being legally able to drink"
Introduction to prediction
So far, we had been focusing on causal inference:
- Estimating an effect and "predicting" a counterfactual (what if?)
Now, we will focus on prediction:
- Estimate/predict outcomes under specific conditions.
Differences between inference and prediction
Inference → focus on covariate
- Interpretability of model.
Prediction → focus on outcome variable
- Accuracy of model.
Both can be complementary!
Example: What is churn?
- Churn: Measure of how many customers stop using your product (e.g. cancel a subscription).
Example: What is churn?
- Churn: Measure of how many customers stop using your product (e.g. cancel a subscription).
Less costly to keep a customer than bring a new one

Example: What is churn?
- Churn: Measure of how many customers stop using your product (e.g. cancel a subscription).
Less costly to keep a customer than bring a new one
Prevent churn

Example: What is churn?
- Churn: Measure of how many customers stop using your product (e.g. cancel a subscription).
Less costly to keep a customer than bring a new one
Prevent churn
Identify customer that are likely to cancel/quit/fail to renew

Bias vs Variance
"There are no free lunches in statistics"
Bias vs Variance
"There are no free lunches in statistics"
Not one method dominates others: Context/dataset dependent.
Remember that the goal of prediction is to have a method that is accurate in predicting outcomes on previously unseen data.
- Validation set approach: Training and testing data
Bias vs Variance
"There are no free lunches in statistics"
Not one method dominates others: Context/dataset dependent.
Remember that the goal of prediction is to have a method that is accurate in predicting outcomes on previously unseen data.
- Validation set approach: Training and testing data
Balance between flexibility and accuracy
Bias vs Variance
Variance
"[T]he amount by which the function f would change if we estimated it using a different training dataset"
Bias vs Variance
Variance
"[T]he amount by which the function f would change if we estimated it using a different training dataset"
Bias
"[E]rror introduced by approximating a real-life problem with a model"
Q1:Which models do you think are higher variance?
a) More flexible models
b) Less flexible models
Bias vs. Variance: The ultimate battle
In inference, bias >> variance
In prediction, we care about both:
- Measures of accuracy will have both bias and variance.
Trade-off at different rates
How do we measure accuracy?
Different measures (for continuous outcomes):
- Remember Adj−R2?
How do we measure accuracy?
Different measures (for continuous outcomes):
Remember Adj−R2?
- R2 (proportion of the variation in Y explained by Xs) adjusted by the number of predictors!
How do we measure accuracy?
Different measures (for continuous outcomes):
Remember Adj−R2?
- R2 (proportion of the variation in Y explained by Xs) adjusted by the number of predictors!
Mean Squared Error (MSE): Can be decomposed into variance and bias terms MSE=1nn∑i=1(yi−^f(xi))2
How do we measure accuracy?
Different measures (for continuous outcomes):
Remember Adj−R2?
- R2 (proportion of the variation in Y explained by Xs) adjusted by the number of predictors!
Mean Squared Error (MSE): Can be decomposed into variance and bias terms MSE=1nn∑i=1(yi−^f(xi))2
Root Mean Squared Error (RMSE): Measured in the same units as the outcome! RMSE=√MSE
How do we measure accuracy?
Different measures (for continuous outcomes):
Remember Adj−R2?
- R2 (proportion of the variation in Y explained by Xs) adjusted by the number of predictors!
Mean Squared Error (MSE): Can be decomposed into variance and bias terms MSE=1nn∑i=1(yi−^f(xi))2
Root Mean Squared Error (RMSE): Measured in the same units as the outcome! RMSE=√MSE
Other measures: Bayesian Information Criterion (BIC) and Akaike Information Criterion (AIC)
Is flexibility always better?
Is flexibility always better?
Is flexibility always better?
Is flexibility always better?
Example: Let's predict "pre-churn"!
- You work at HBO Max and you know that a good measure for someone at risk of unsubscribing is the times they've logged in the past week:
hbo = read.csv("https://raw.githubusercontent.com/maibennett/sta235/main/exampleSite/content/Classes/Week10/1_ModelSelection/data/hbomax.csv")head(hbo)## id female city age logins succession unsubscribe## 1 1 1 1 53 10 0 1## 2 2 1 1 48 7 1 0## 3 3 0 1 45 7 1 0## 4 4 1 1 51 5 1 0## 5 5 1 1 45 10 0 0## 6 6 1 0 40 0 1 0Two candidates: Simple vs Complex
Simple Model:
logins=β0+β1×Succession+β2×city+ε
Complex Model:
logins=β0+β1×Succession+β2×age+β3×age2+β4×city+β5×female+ε
Create Validation Sets
set.seed(100) #Always set seed for replication!n = nrow(hbo)train = sample(1:n, n*0.8) #randomly select 80% of the rows for our training sampletrain.data = hbo %>% slice(train)test.data = hbo %>% slice(-train)Create Validation Sets
set.seed(100) #Always set seed for replication!n = nrow(hbo)train = sample(1:n, n*0.8)train.data = hbo %>% slice(train)test.data = hbo %>% slice(-train)Create Validation Sets
set.seed(100) #Always set seed for replication!n = nrow(hbo)train = sample(1:n, n*0.8) #randomly select 80% of the rows for our training sampletrain.data = hbo %>% slice(train)test.data = hbo %>% slice(-train)Estimate Accuracy Measure
library(modelr)lm_simple = lm(logins ~ succession + city, data = train.data)lm_complex = lm(logins ~ female + city + age + I(age^2) + succession, data = train.data)# For simple model:rmse(lm_simple, test.data) %>% round(., 4)## [1] 2.0899# For complex model:rmse(lm_complex, test.data) %>% round(., 4)## [1] 2.0934- Q2: Which one would you prefer?
Cross-Validation
- To avoid using only one training and testing dataset, we can iterate over k-fold division of our data:
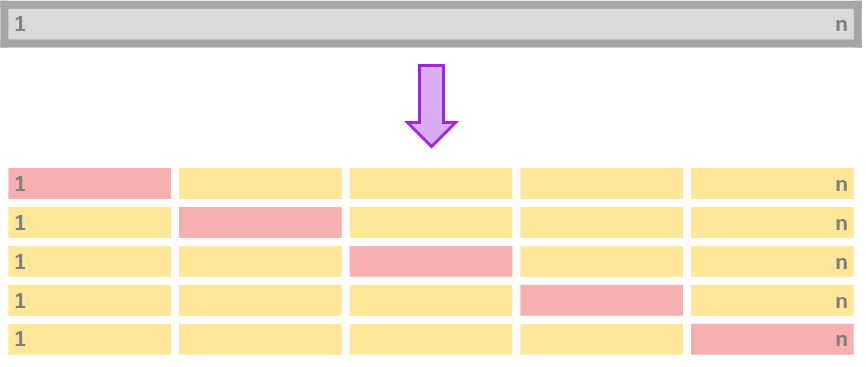
Cross-Validation
Procedure for k-fold cross-validation:
Divide your data in k-folds (usually, K=5 or K=10).
Use k=1 as the testing data and k=2,..,K as the training data.
Calculate the accuracy measure Ak on the testing data.
Repeat for each k.
Average Ak for all k∈K.
Cross-Validation
Procedure for k-fold cross-validation:
Divide your data in k-folds (usually, K=5 or K=10).
Use k=1 as the testing data and k=2,..,K as the training data.
Calculate the accuracy measure Ak on the testing data.
Repeat for each k.
Average Ak for all k∈K.
Main advantage: Use the entire dataset for training AND testing.
How do we do CV in R?
library(caret)set.seed(100)train.control = trainControl(method = "cv", number = 10)lm_simple = train(logins ~ succession + city, data = disney, method="lm", trControl = train.control)lm_simpleHow do we do CV in R?
library(caret) set.seed(100)train.control = trainControl(method = "cv", number = 10)lm_simple = train(logins ~ succession + city, data = disney, method="lm", trControl = train.control)lm_simpleHow do we do CV in R?
library(caret) set.seed(100)train.control = trainControl(method = "cv", number = 10)lm_simple = train(logins ~ succession + city, data = disney, method="lm", trControl = train.control)lm_simpleHow do we do CV in R?
library(caret) set.seed(100)train.control = trainControl(method = "cv", number = 10)lm_simple = train(logins ~ succession + city, data = hbo, method="lm", trControl = train.control)lm_simple## Linear Regression ## ## 5000 samples## 2 predictor## ## No pre-processing## Resampling: Cross-Validated (10 fold) ## Summary of sample sizes: 4500, 4501, 4499, 4500, 4500, 4501, ... ## Resampling results:## ## RMSE Rsquared MAE ## 2.087314 0.6724741 1.639618## ## Tuning parameter 'intercept' was held constant at a value of TRUEStepwise selection
We have seen how to choose between some given models. But what if we want to test all possible models?
Stepwise selection: Computationally-efficient algorithm to select a model based on the data we have (subset selection).
Stepwise selection
We have seen how to choose between some given models. But what if we want to test all possible models?
Stepwise selection: Computationally-efficient algorithm to select a model based on the data we have (subset selection).
Algorithm for forward stepwise selection:
Start with the null model, M0 (no predictors)
For k=0,...,p−1: (a) Consider all p−k models that augment Mk with one additional predictor. (b) Choose the best among these p−k models and call it Mk+1.
Select the single best model from M0,...,Mp using CV.
Backwards stepwise follows the same procedure, but starts with the full model.
Will forward stepwise subsetting yield the same results as backwards stepwise selection?
How do we do stepwise selection in R?
set.seed(100)train.control = trainControl(method = "cv", number = 10) #set up a 10-fold cvlm.fwd = train(logins ~ . - unsubscribe, data = train.data, method = "leapForward", tuneGrid = data.frame(nvmax = 1:5), trControl = train.control)lm.fwd$results## nvmax RMSE Rsquared MAE RMSESD RsquaredSD MAESD## 1 1 2.269469 0.6101788 1.850376 0.04630907 0.01985045 0.04266950## 2 2 2.087184 0.6702660 1.639885 0.04260047 0.01784601 0.04623508## 3 3 2.087347 0.6702094 1.640405 0.04258030 0.01804773 0.04605074## 4 4 2.088230 0.6699245 1.641402 0.04270561 0.01808685 0.04620206## 5 5 2.088426 0.6698623 1.641528 0.04276883 0.01810569 0.04624618How do we do stepwise selection in R?
set.seed(100)train.control = trainControl(method = "cv", number = 10) #set up a 10-fold cvlm.fwd = train(logins ~ . - unsubscribe, data = train.data, method = "leapForward", tuneGrid = data.frame(nvmax = 1:5), trControl = train.control)lm.fwd$results## nvmax RMSE Rsquared MAE RMSESD RsquaredSD MAESD## 1 1 2.269469 0.6101788 1.850376 0.04630907 0.01985045 0.04266950## 2 2 2.087184 0.6702660 1.639885 0.04260047 0.01784601 0.04623508## 3 3 2.087347 0.6702094 1.640405 0.04258030 0.01804773 0.04605074## 4 4 2.088230 0.6699245 1.641402 0.04270561 0.01808685 0.04620206## 5 5 2.088426 0.6698623 1.641528 0.04276883 0.01810569 0.04624618- Which one would you choose out of the 5 models? Why?
How do we do stepwise selection in R?
# We can see the number of covariates that is optimal to choose:lm.fwd$bestTune## nvmax## 2 2# And how does that model looks like:summary(lm.fwd$finalModel)## Subset selection object## 5 Variables (and intercept)## Forced in Forced out## id FALSE FALSE## female FALSE FALSE## city FALSE FALSE## age FALSE FALSE## succession FALSE FALSE## 1 subsets of each size up to 2## Selection Algorithm: forward## id female city age succession## 1 ( 1 ) " " " " " " " " "*" ## 2 ( 1 ) " " " " "*" " " "*"# If we want the RMSErmse(lm.fwd, test.data)## [1] 2.089868Your Turn
Takeaway points
In prediction, everything is going to be about bias vs variance.
Importance of validation sets.
We now have methods to select models.

Next class
Continue with prediction and model selection
Shrinkage/Regularization methods:
- Ridge regression and Lasso.

References
James, G. et al. (2021). "Introduction to Statistical Learning with Applications in R". Springer. Chapter 2, 5, and 6.
STDHA. (2018). "Stepwise Regression Essentials in R."
STDHA. (2018). "Cross-Validation Essentials in R."